


| Upadated, more comprehensive descriptions are coming soon... |
|
(1) Simulated fibers driven by a 3D-Fractional Brownian Motion in the superdiffusion regime (H = 0.85). This model closely approximates the behavior or serotonergic fibers in mammalian brains. (2) Simulated fibers driven by a 3D-Fractional Brownian Motion in the subdiffusion regime (H = 0.35). For details, see Janusonis et al. (2020). Virtually all neural processes in the mammalian brain are embedded in a matrix of meandering fibers that release serotonin, a signaling molecule that dates back to the earliest multicellular life forms. The density of this "serotonergic" matrix varies across brain regions, and its perturbations can profoundly affect perception, emotional states, and transitions between wakefulness and sleep. In humans, altered densities of serotonin-releasing fibers have been associated with Autism Spectrum Disorder, prolonged exposure to MDMA ("Ecstasy"), and other conditions. It is currently assumed that brain regions use an unknown mechanism to actively build and maintain specific serotonergic fiber densities. We have introduced an alternative and more parsimonious hypothesis that states that region-specific densities arise as a mathematical consequence of the stochastic behavior of individual fibers, with no global supervision. In the project, we investigate the stochastic processes that drive single serotonergic fibers. This collaborative effort brings together experimental neurobiology, advanced mathematics, and supercomputing. |
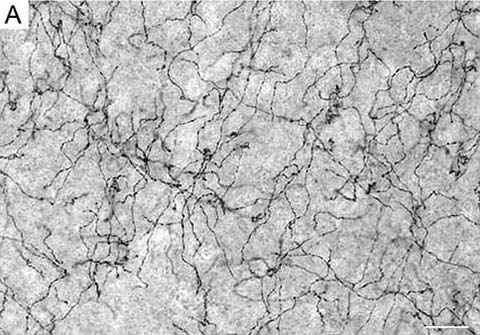
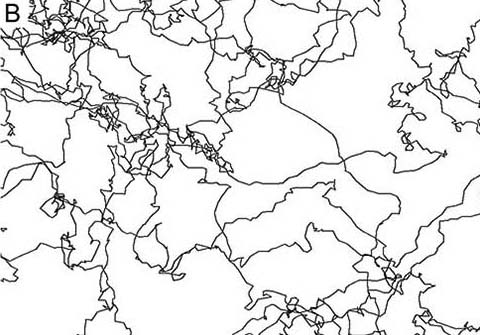
(A) Serotonergic fibers in the mouse cerebral cortex. Scale bar = 20 µm. (B) Simulated fibers driven by Fractional Brownian Motion in the superdiffusion regime. For details, see Janusonis et al. (2020).
 This research has been supported by the National Science Foundation, the National Institute of Mental Health, and the California NanoSystems Institute |
|
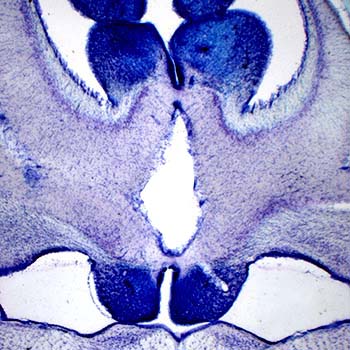
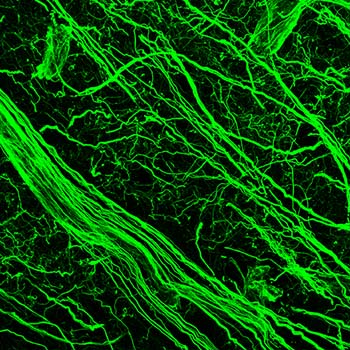
(1) A section through a shark cerebellum. (2) Iba1-positive ependymoglia in the swellshark forebrain. Many elasmobranchs (sharks, skates, rays) have highly evolved brains that conform to the general vertebrate architecture. However, they also show an extreme variability in how brain divisions are built anatomically and used functionally. This rich set of possibilities reveals which mammalian brain features are highly idiosyncratic and represent only one realization among many others. It also demonstrates what future brains may still become, by evolution or human design. We study the brain cytoarchitecture of a number of shark species, in both the Galeomorphii and Squalomorphii superorders. Some of the extant clades date back to the Triassic (pre-Jurassic) period. We have recently discovered that swellsharks express Iba1 (a mammalian microglia-specific protein) in radial ependymoglia, a major element of the shark brain scaffolding. We also investigate serotonergic fibers in this clade.  This research has been supported by the National Science Foundation |